GpOptimiser
- class inference.gp.GpOptimiser(x: ~numpy.ndarray, y: ~numpy.ndarray, bounds: ~collections.abc.Sequence, y_err: ~numpy.ndarray = None, hyperpars: ~numpy.ndarray = None, kernel: ~inference.gp.covariance.CovarianceFunction = <class 'inference.gp.covariance.SquaredExponential'>, mean: ~inference.gp.mean.MeanFunction = <class 'inference.gp.mean.ConstantMean'>, cross_val: bool = False, acquisition=<class 'inference.gp.acquisition.ExpectedImprovement'>, optimizer: str = 'bfgs', n_processes: int = 1)
A class for performing Gaussian-process optimisation in one or more dimensions.
GpOptimiser extends the functionality of GpRegressor to perform Gaussian-process optimisation, often also referred to as ‘Bayesian optimisation’. This technique is suited to problems for which a single evaluation of the function being explored is expensive, such that the total number of function evaluations must be made as small as possible.
In order to construct the Gaussian-process regression estimate which is used to search for the global maximum, on initialisation GpOptimiser must be provided with at least two evaluations of the function which is to be maximised.
- Parameters
x – The x-data points as a 2D
numpy.ndarraywith shape (number of points, number of dimensions). Alternatively, a list of array-like objects can be given, which will be converted to andarrayinternally.y – The y-data values as a 1D
numpy.ndarray.bounds – An iterable containing tuples which specify the upper and lower bounds for the optimisation in each dimension in the format (lower_bound, upper_bound).
y_err – The error on the y-data values supplied as a 1D array. This technique explicitly assumes that errors are Gaussian, so the supplied error values represent normal distribution standard deviations. If this argument is not specified the errors are taken to be small but non-zero.
hyperpars – An array specifying the hyper-parameter values to be used by the covariance function class, which by default is
SquaredExponential. See the documentation for the relevant covariance function class for a description of the required hyper-parameters. Generally this argument should be left unspecified, in which case the hyper-parameters will be selected automatically.kernel (class) – The covariance-function class which will be used to model the data. The covariance-function classes can be imported from the
gpmodule and then passed toGpOptimiserusing this keyword argument.mean (class) – The mean-function class which will be used to model the data. The mean-function classes can be imported from the
gpmodule and then passed toGpOptimiserusing this keyword argument.cross_val (bool) – If set to
True, leave-one-out cross-validation is used to select the hyper-parameters in place of the marginal likelihood.acquisition (class) – The acquisition-function class which is used to select new points at which the objective function is evaluated. The acquisition-function classes can be imported from the
gpmodule and then passed as arguments - see their documentation for the list of available acquisition functions. If left unspecified, theExpectedImprovementacquisition function is used by default.optimizer (str) – Selects the optimisation method used for selecting hyper-parameter values and proposed evaluations. Available options are “bfgs” for
scipy.optimize.fmin_l_bfgs_bor “diffev” forscipy.optimize.differential_evolution.n_processes (int) – Sets the number of processes used when selecting hyper-parameters or proposed evaluations. Multiple processes are only used when the optimizer keyword is set to “bfgs”.
- add_evaluation(new_x: ndarray, new_y: ndarray, new_y_err: ndarray = None)
Add the latest evaluation to the data set and re-build the Gaussian process so a new proposed evaluation can be made.
- Parameters
new_x – location of the new evaluation
new_y – function value of the new evaluation
new_y_err – Error of the new evaluation.
- propose_evaluation(optimizer=None)
Request a proposed location for the next evaluation. This proposal is selected by maximising the chosen acquisition function.
- Parameters
optimizer (str) – Selects the optimization method used for selecting the proposed evaluation. Available options are “bfgs” for
scipy.optimize.fmin_l_bfgs_bor “diffev” forscipy.optimize.differential_evolution. This keyword allows the user to override the choice of optimizer given whenGpOptimiserwas initialised.- Returns
location of the next proposed evaluation.
Example code
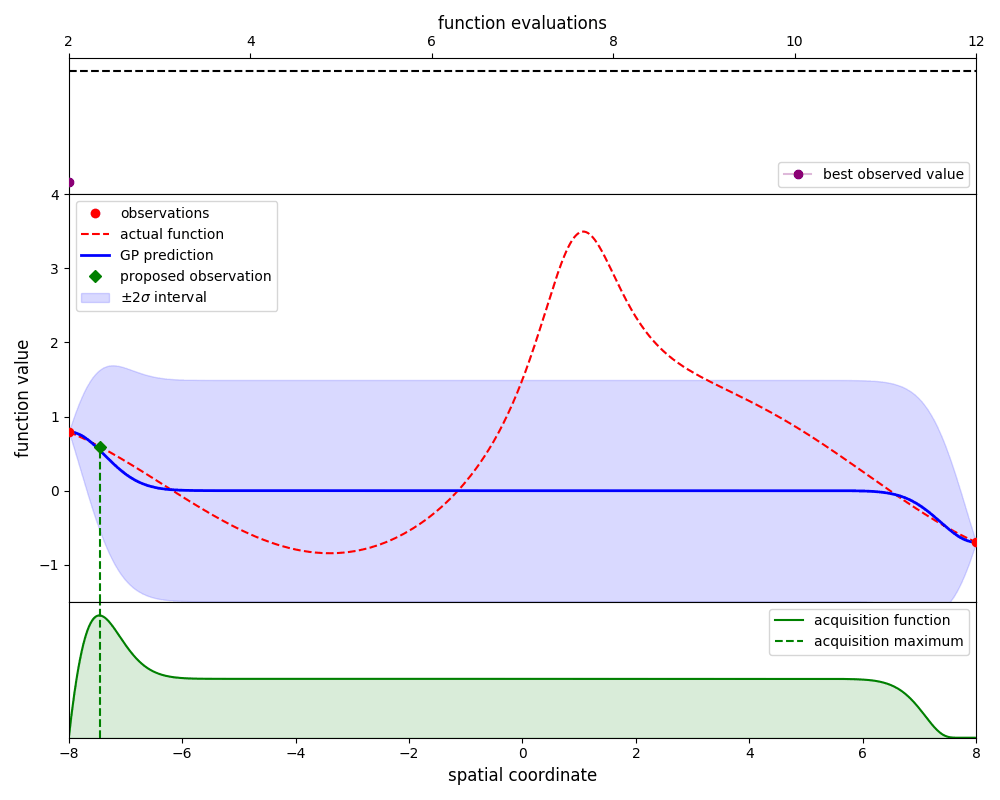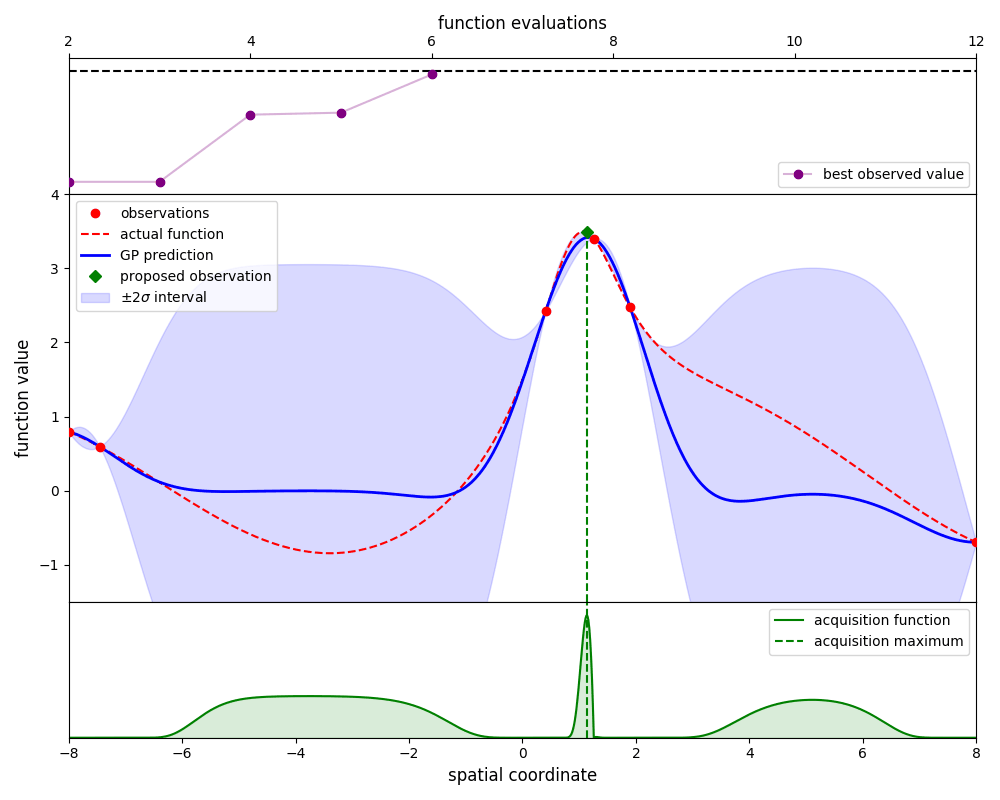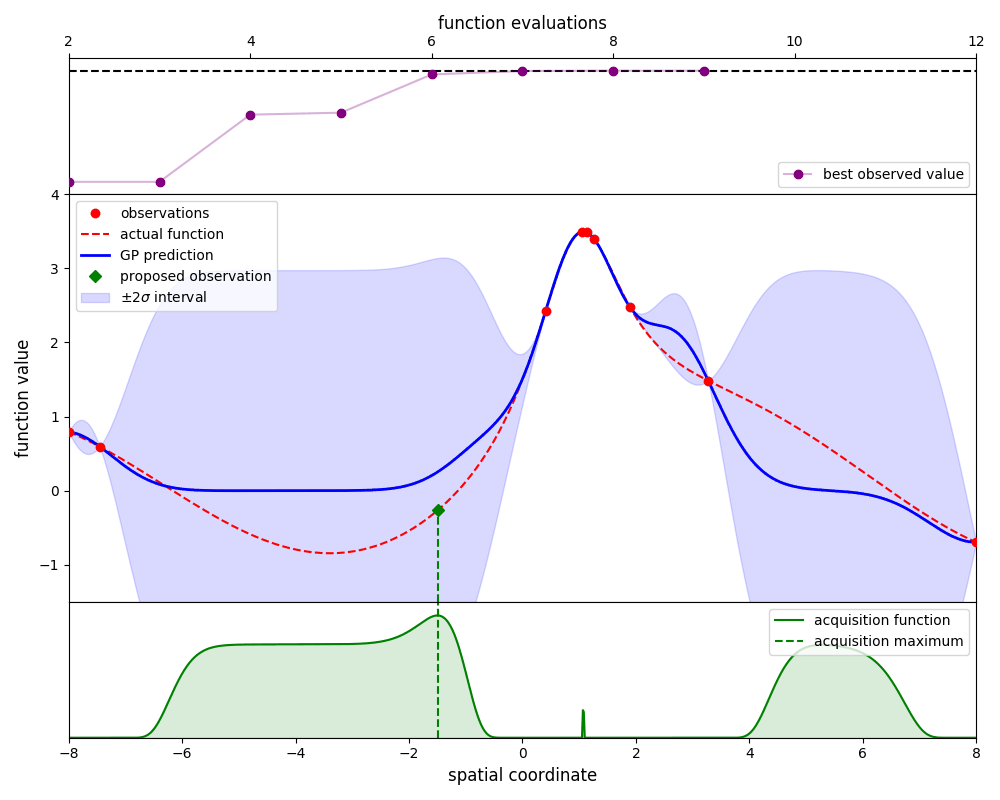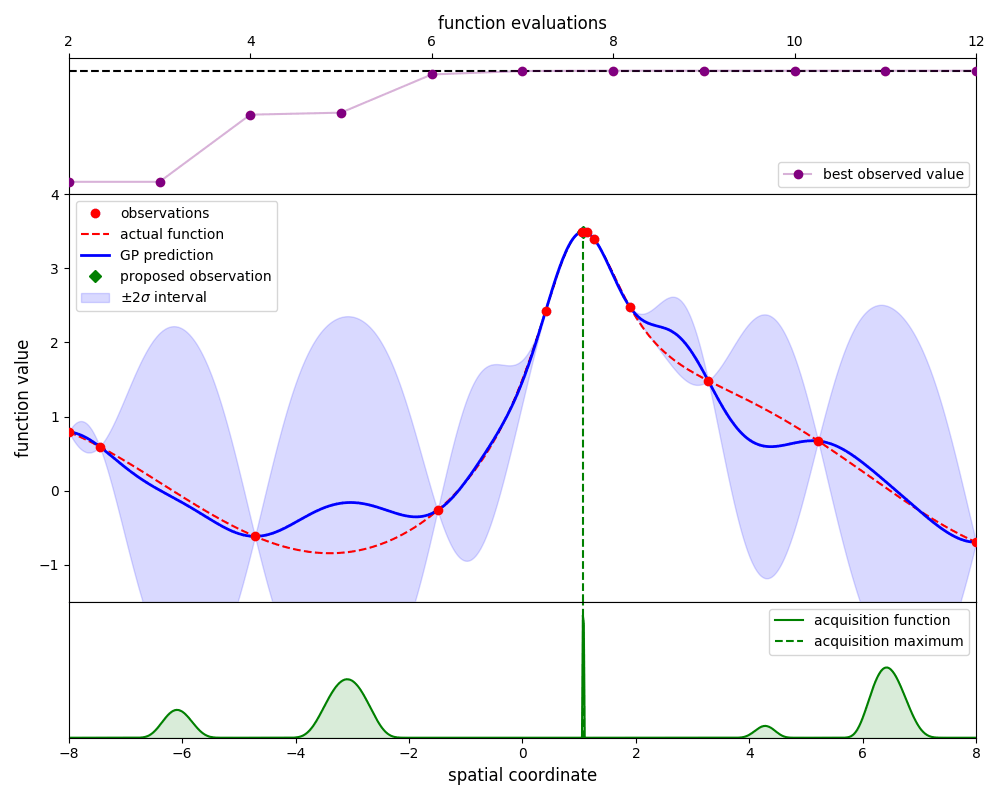
Gaussian-process optimisation efficiently searches for the global maximum of a function by iteratively ‘learning’ the structure of that function as new evaluations are made.
As an example, define a simple 1D function:
from numpy import sin
def search_function(x): # Lorentzian plus a sine wave
return sin(0.5 * x) + 3 / (1 + (x - 1)**2)
Define some bounds for the optimisation, and make some evaluations of the function that will be used to build the initial gaussian-process estimate:
# define bounds for the optimisation
bounds = [(-8.0, 8.0)]
# create some initialisation data
x = array([-8.0, 8.0])
y = search_function(x)
Create an instance of GpOptimiser:
from inference.gp import GpOptimiser
GP = GpOptimiser(x, y, bounds=bounds)
By using the propose_evaluation method, GpOptimiser will propose a new evaluation of
the function. This proposed evaluation is generated by maximising an acquisition function,
in this case the ‘expected improvement’ function. The new evaluation can be used to update
the estimate by using the add_evaluation method, which leads to the following loop:
for i in range(11):
# request the proposed evaluation
new_x = GP.propose_evaluation()
# evaluate the new point
new_y = search_function(new_x)
# update the gaussian process with the new information
GP.add_evaluation(new_x, new_y)
Here we plot the state of the estimate at each iteration: